Making precision oncology more personal
FROM GENETIC TESTING FOR CANCER TO FUNCTIONAL, PATIENT-DERIVED MODELS
Genetic testing for cancer has rapidly become an important and routine part of oncology treatment. It’s driven the rise of precision oncology, where clinicians seek to match the right patient and their genetic profile, with the right drug.1,2 The detection of genetic biomarkers using next-generation sequencing (NGS) or other genetic techniques has been foundational for precision oncology these two concepts are now considered nearly interchangeable. Even the National Cancer Institute equates precision medicine with the underlying genetics of a disease like cancer with statements like: “Precision medicine is an approach to patient care that allows doctors to select treatments that are most likely to help patients based on a genetic understanding of their disease.”

It’s easy to see why drug developers, clinicians, patients and caregivers have rapidly adopted this view. The idea of doing a fast and simple genetic test and identifying safe, efficacious and targeted therapies brings hope and promise to those processing a recent diagnosis, living with a life-changing disease, or making a decision about a first-, second-, or third-line treatment regimen.
While this concept is both comforting and enchanting, in reality, genetic testing hasn’t delivered on the hope and promise of precision oncology and the majority of patients experience disappointing clinical outcomes.1 With the limits of this “go-to” technique quickly being realized, the time for new, personalized approaches has come.
Let’s take a closer look at the issues with genetic testing and dive into the promise of more precise and personalized methodologies.
The current state of precision medicine and cancer: An incomplete picture
Cancer is regarded as primarily a genetic disease and so it makes sense why genomic testing has been used as the basis for treatment decisions.3 And there have certainly been some incredible successes in precision oncology: Gleevac treatment of chronic myeloid leukemia with BCR-ABL fusion kinase, epidermal growth factor receptor (EGFR) inhibitor treatment of lung cancers with EGFR mutations, and BRAF inhibitor treatment of melanomas with BRAF mutations have all proven successful.1 There are also countless anecdotes of cancer patients benefiting from genome-guided therapies.
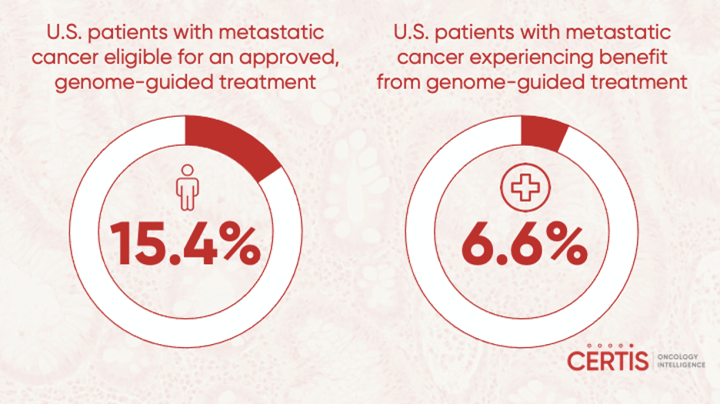
Yet, genetics doesn’t tell the whole story, and using genomic testing to guide treatment selection only benefits a minority of patients.1 The proportion of patients whose treatment decisions can be chosen through the detection of biomarkers is estimated to be between 10% and 50%.4 In 2018, an analysis published in JAMA Oncology reported that only 15.4% of U.S. patients with metastatic cancer were eligible for an approved, genome-guided drug but only a small percentage, 6.6% of those that were treated experienced a clinical benefit.5 The reasons for the low response are multifaceted.
Driving vs. passenger mutations
For one, the genetic complexity of cancer and the complicated interplay between specific mutations and their role in tumor biology are major underlying issues. Targeted NGS panels may look at the most common oncogenic mutations, but there is no way of telling how these mutations are driving cancer progression. The mere presence of a mutation doesn’t tell clinicians about its role in a patient’s disease or how a patient will respond to targeted therapies. Some mutations may simply be 'passengers' that don’t contribute to the progression of a patient’s cancer, and genomic testing has no way to distinguish between relevant versus irrelevant mutations.6
Furthermore, if a driving mutation is correctly identified, there is no guarantee that there is a targeted therapeutic available for treatment. One major requirement for the identification of safe and efficacious treatments is a large body of scientific evidence correlating a driving mutation with response to a specific treatment. The absence of relevant or convincing evidence can make it difficult for patients and clinicians to confidently select a treatment, even with genomic data.
Insufficient genomic coverage and depth
In addition, targeted NGS techniques can have sequencing errors and may not have the capability of identifying every single mutation within a patient’s genome. So oncogenic or resistance mutations that make a genome-guided treatment ineffective may be missed during genetic testing, offering an incomplete picture on which to base treatment decisions. For instance, mutations in PTEN make lung cancers with specific EGFR mutations resistant to EGFR inhibitors.7
Genetic testing is a static snapshot
Finally, genetic testing doesn’t take into account the accumulation of additional 'driver' or 'passenger' mutations over time. It offers a static genetic picture, frozen in time. But a patient’s cancer is likely to evolve, accumulating new mutations that may lead to a change in clinical course or reduce the efficacy of a treatment.
Taken together, these ambiguities and complicating factors can make it difficult for oncologists to confidently select treatments, based on targeted sequencing panels. With these shortcomings, it may seem that using genetic testing to inform cancer treatment is a bit like trying to pick the fastest path to a destination with part of your map missing or trying to assemble a piece of office furniture with steps three through 10 missing from your instructions.
Thankfully, other techniques are taking a more personalized approach with tumor cells derived directly from a patient.
Moving beyond genomic testing
These methods provide a much-needed alternative to genomic testing and can act as a natural complement. Some clinicians may recommend throwing the precision oncology baby out with the bathwater, but other oncology leaders offer a more refined and measured path forward. In a unifying Perspective article published in Nature Medicine, Anthony Letai, of the Dana-Farber Cancer Institute, explains his argument for developing an expanded precision oncology toolkit: “Instead of relying exclusively on big data measurements of initial conditions, we should also acquire highly actionable functional information by perturbing – for example, with cancer therapies – viable primary tumor cells from patients with cancer.”1 He continues with his argument that genomics is useful in precision oncology and can help in “...elucidating mechanisms of tumorigenesis, documenting intratumoral heterogeneity, and detecting clonal evolution in therapy, even when it is not useful in assigning therapies to patients.”1

In using patient-derived tumors, clinicians and patients can measure the most clinically relevant outcomes – treatment response – in the most personalized way possible. There are several different approaches (with advantages and drawbacks) to this functional version of precision oncology, and while few of them have been applied in clinical trials, they hold incredible promise, and have been validated in both academic and private studies.
Culturing patient-derived tumors in a 2D environment
Perhaps the simplest way of functionally testing patient-derived tumors is by culturing them ex vivo in a 2D monolayer and exposing them to potential therapeutics. While simple, fast and inexpensive, these techniques fall short of mimicking the complex 3D tumor microenvironment or taking into account the complex interplay between tumor and host cells.1 Because this technique is ex vivo, it also doesn’t allow researchers to track other clinical characteristics, such as metastasis.
Nevertheless, this technique has proven somewhat successful in identifying possible treatment options for patients with various types of leukemia.1 The technique has yet to be applied to solid tumors, but is being tested in prospective clinical trials for acute myelogenous leukemia (AML), acute lymphoblastic leukemia (ALL), and chronic lymphocytic leukemia (CLL).1
Culturing patient-derived tumors in a 3D environment
An emerging technique that has proven useful in the analysis of patient-derived tumors is the use of tumor organoids. By culturing primary tumor cells in a 3D environment, organoids can more closely recapitulate the morphological and genetic features of a patient’s tumor.1
It also allows researchers to more accurately mimic the tumor microenvironment through the inclusion of host cells from the original biopsy or surgical specimen. These models can then be used for treatment testing, and primary tumors from the breast, colon, pancreas and prostate have been used to generate organoids.8 There is also mounting evidence that observed in vitro drug sensitivity holds true in the clinic.1
Patient-derived xenografts (PDX) Models
PDX models are a natural step closer to mimicking the exact tumor microenvironment that might occur in a patient. Instead of culturing a patient’s tumor ex vivo, tumors are implanted into immunodeficient mouse models, passaged and subjected to pharmacological testing to identify treatments that may be clinically relevant.1
The advantages of using this method are numerous and include a clinically accurate tumor growth, treatment response, recurrence and acquired drug resistance. Some PDX techniques are better at replicating clinically relevant tumor characteristics than others. For instance, orthotopic PDX (O-PDX) models can mimic and predict tumor metastasis whereas subcutaneous methods may not.9 It had been previously reported that these models didn’t accurately recapitulate the in vivo genetics of a patient’s tumor, but that has recently been refuted by an international research group that included the PDXNET Consortium, and EurOPDX Consortium.10
There are some disadvantages to PDX models, including length of time (a scale of multiple months) and amount of money required to establish them.1,8 However, there is mounting evidence that PDX models, specifically O-PDX, may provide a more precise and more personalized way of analyzing a patient’s tumor and the treatments it will respond to in the clinic.

Precision oncology is Evolving
The emergence of new functional precision oncology techniques is helping patients and clinicians make better treatment decisions. In addition, these new techniques are helping drug developers create new targeted therapies to drive better patient outcomes. With additional clinical testing and “co-clinical” trials, these techniques can be used side-by-side genomic testing. And with this additional data and insight, a clearer picture of what precision oncology can do for patients and the pharmaceutical industry will snap into focus.
Certis has helped patients and their care teams make better treatment decisions. Learn more about the O-PDX model approach with these resources:
- Webinar: Testing sarcoma drug response in personalized mouse avatars
- Publications: O-PDX
- FAQs: O-PDX for drug developers
References
- Letai A. Functional precision cancer medicine—moving beyond pure genomics. Nat Med. 2017; 23: 1028–1035 (2017).
- Precision medicine in cancer treatment. National Cancer Institute website: Published Oct. 3, 2017. Accessed April 27, 2021.
- The Genetics of Cancer. National Cancer Institute website: Published October 12, 2017. Accessed April 27, 2021.
- Beaubier N, Bontrager M, Huether R, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat Biotechnol. 2019; 37:1351–1360.
- A cancer drug tailored to your tumor? Experts trade barbs over ‘precision oncology’. Science website: Published April 24, 2018. Accessed April 27, 2021.
- Wodarz D, Newell AC, Komarova NL. Passenger mutations can accelerate tumour suppressor gene inactivation in cancer evolution. J R Soc Interface. 2018; 15(143): 20170967.
- Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR [published correction appears in Cancer Res. 2015 May 1; 75(9): 1922]. Cancer Res. 2009; 69(8): 3256-3261.
- Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020; 13(1):4.
- Bhimani J, Ball K, Stebbing J. Patient-derived xenograft models—the future of personalised cancer treatment. Br J Cancer. 2020; 122: 601–602.
- Woo XY, Giordano J, Srivastava A, et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nature Genetics. 2021 Jan; 53(1): 86-99.
About the Author:
Elie Diner has a Ph.D. in bioengineering and 12 years of research experience in microbiology, synthetic biology and immunology. During his time at the bench, he developed a passion for effective science communication and eventually transitioned into a career as a professional science and content writer. He's authored 12 peer-reviewed scientific publications and numerous blogs, whitepapers, and e-books for life science companies.
Back to Feed